Cervical precancerous lesions are curable when detected early, and visual interpretation via histopathology slides is an important criterion during diagnosis. However, only professional pathologists can accurately determine cervical precancerous lesions through histopathological images, because of the complexity of the cervical tissue structure and the diversity of cell morphology. MTCHI is a public dataset for the exploration of cervical precancerous lesions to help computer experts without medical background to delve and compare the automated algorithms. The data in the MTCHI dataset are provided by Singularity.AI Technology.
Two preliminary tasks are designed based on the characteristics and clinical needs of the cervical histopathology images :
(1) rapid extraction of foreground tissue objects (pre-processing),
(2) segmentation of precancerous lesions in the regions of interest (RoIs).
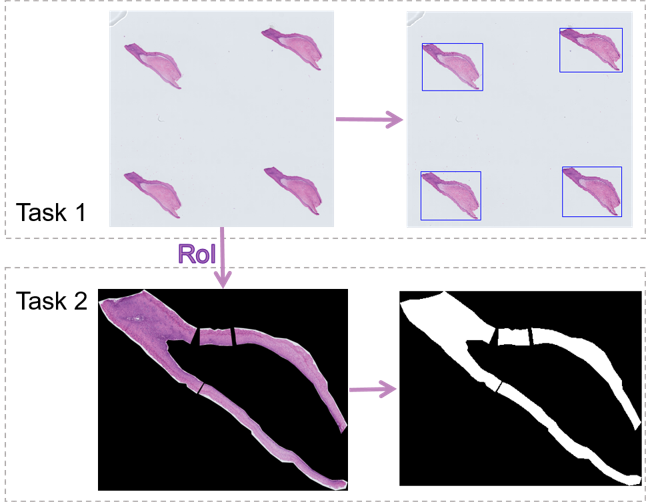
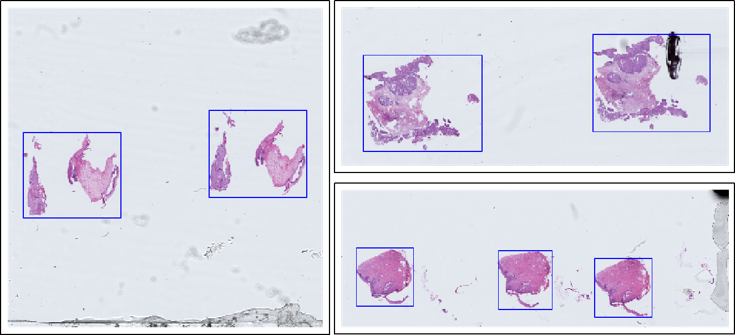
Task 1 is designed as the preprocessing for machine learning and tool for pathologists to reduce the time burden. Considering the difficulty of annotation and the requirements of the actual application, the foreground objects are annotated with rectangular bounding boxes. Task 1 does not set up a training set and all images are used for testing, thereby allowing the exploration of a common fast foreground extraction algorithm.
Task 2 aims at pixel-level segmentation of cervical precancerous lesions. The RoIs are selected by pathologists because of the complication of the cervical tissue structure. The data for Task 2 are obtained by cropping the images surrounding the RoIs. The regions outside the RoIs are regarded as the background, that is, all the three channels (RGB) of every pixel are set to zero. The RoIs are annotated pixel by pixel into four categories, namely, Normal, CIN1, CIN2, and CIN3.
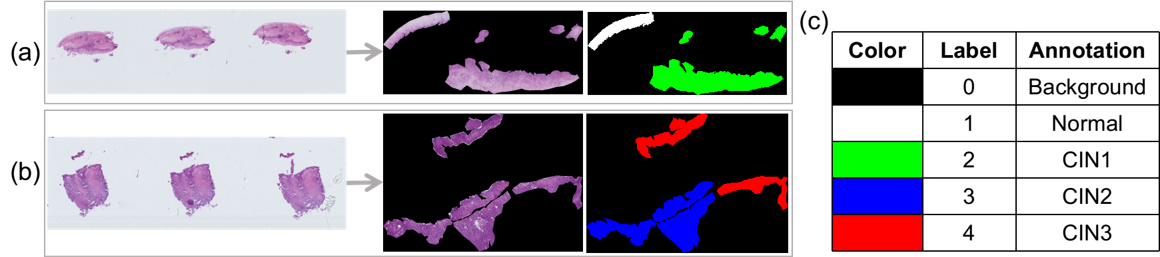
Additional data:
Pixel-level labeling is laborious and time-consuming, while patient-level annotations are relatively abundant in hospitals. Thus, using massive unlabeled data to improve the performance of the algorithms is highly encouraged. To facilitate algorithm exploration and comparison, additional 80 cervical histopathological images with image-level annotations are provided. The data are annotated as multi-labels by two independent pathologists. For example, an image is annotated as “CIN 1 and CIN 2”, if both lesions can be found in the image with corresponding regions.
Please Cite:
[1] Z. Meng, Z. Zhao, B. Li, F. Su and L. Guo, "A Cervical Histopathology Dataset for Computer Aided Diagnosis of Precancerous Lesions," in IEEE Transactions on Medical Imaging, doi: 10.1109/TMI.2021.3059699.
[2] Z. Meng, Z. Zhao, B. Li, F. Su, L. Guo and H. Wang, "Triple Up-sampling Segmentation Network with Distribution Consistency Loss for Pathological Diagnosis of Cervical Precancerous Lesions," in IEEE Journal of Biomedical and Health Informatics, doi: 10.1109/JBHI.2020.3043589.